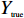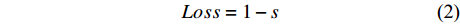Prediction of Response to Neoadjuvant Chemotherapy for Breast Cancer Based on Histomorphology Analysis of Needle Biopsy Images
-
摘要:目的 利用深度学习的方法对乳腺癌患者接受新辅助化疗(NAC)前的穿刺切片进行肿瘤区域和细胞核的自动分割,提取肿瘤区域细胞群特征,从而对乳腺癌NAC病理缓解程度进行预测。方法 收集在江苏省人民医院接受NAC治疗前的68位乳腺癌患者的术前穿刺HE染色切片,两位病理医生对其中12张穿刺切片进行了肿瘤区域的标记,其中8张作为训练集,4张作为测试集,剩余的56张由训练好的肿瘤区分割模型进行肿瘤分割。运用UNet++建立分割模型,分别对乳腺癌病理穿刺切片肿瘤区域和细胞核进行自动分割;然后,根据自动分割的肿瘤区域内细胞核,构建肿瘤内细胞层次的特征;最后运用特征选择方法选择有效的特征,通过五折交叉验证训练分类器模型预测NAC的病理缓解程度的高低。结果 基于68位患者的病理穿刺切片进行预测,最大相关最小冗余(mRMR)的特征选择方法筛选出的10个维度特征和随机森林(RF)分类器结合训练的模型预测结果的准确率最高,准确率达到82.35%,曲线下面积(AUC)值达到0.9082。结论 本模型能够在切片病理图像上自动分割肿瘤区域和细胞核,构建的肿瘤区域细胞核群的特征能够预测患者对NAC的病理缓解程度,方法可靠且可重复性较高,同时发现肿瘤区域细胞核纹理特征在预测中效果较好,进一步证实了肿瘤区域细胞核群对疗效预测具有重要意义。Abstract:Objective The deep learning method was used to automatically segment the tumor area and the cell nucleus based on needle biopsy images of breast cancer patients prior to receiving neoadjuvant chemotherapy (NAC), and then, the features of the cell clusters in the tumor area were identified to predict the level of pathological remission of breast cancer after NAC.Methods 68 breast cancer patients who were to receive NAC at Jiangsu Province Hospital were recruited and the hematoxylin-eosin (HE) stained preoperative biopsy sections of these patients were collected. Unet++ was used to establish a segmentation model and the tumor area and nucleus of the needle biopsy images were automatically segmented accordingly. Then, according to the nuclei in the automatically segmented tumor area, the features of the cells in the tumor were constructed. After that, effective features were selected through the feature selection method and the classifier model was constructed and trained with five-fold cross validation to predict the degree of post-NAC pathological remission.Results Predictions were made based on the needle biopsy images of the 68 patients. The model that combined the 10-dimensional features selected with the minimal redundancy-maximum-relevancy approach (mRMR) and training with the random forest (RF) classifier had the highest prediction accuracy, reaching 82.35%, and an area under curve (AUC) value of 0.9082.Conclusion This model automatically segments tumor areas and cell nucleus on the biopsy images. The features of the cell clusters which are analyzed and identified in the tumor area can be used to predict the pathological response of the patient to NAC. The method is reliable and replicable. In addition, we found that the textural features of cells in the tumor area was a useful predictor of patient response to NAC, which further confirmed that cell cluster in the tumor area is of great significance to the prediction of treatment outcome.
-
乳腺癌是严重威胁女性健康的重要疾病,位居全世界女性癌症发病率的第一位[1],约占女性癌症的30%[2]。新辅助化疗(neoadjuvant chemotherapy,NAC)是目前临床治疗局部晚期乳腺癌的重要手段,它能够减小肿瘤病灶、降低肿瘤分期[3]、消灭微小转移病灶,使更多患者施行保乳手术,提高患者术后生存率和生活质量。然而,只有部分患者在进行NAC后具有较好疗效,能达到病理完全缓解,而对于采用NAC无效的患者,不仅可能产生NAC的副反应,而且延误最佳的治疗时间,导致预后不良,因此在化疗前对患者进行疗效预测至关重要[4]。
肿瘤微环境代表了免疫系统如何对肿瘤做出反应的环境[5],因此基于组织形态学分析肿瘤微环境中潜在的预测性和预后生物标志物受到越来越多的关注。KUMAR等[6]认为,NAC的疗效预测与肿瘤大小和细胞密度存在较大联系。大量临床医学研究者发现淋巴浸润程度越高[7]与NAC后病理完全缓解率的升高显著有关,病理完全缓解率可高达93.7%;文献[8-10]中,经过分析乳腺癌术后全扫描切片发现免疫淋巴细胞核密度是评估乳腺癌NAC病理是否完全缓解的重要因子,免疫淋巴细胞核密度越大,化疗疗效越好;NISHIMURA等[11]提出细胞增殖指数Ki-67在NAC前较高而化疗后显著减少的患者,病理缓解程度较高。在其他癌症预测化疗疗效的类似工作中,文献[12]通过量化分析细胞核特征,如细胞核空间分布、混乱度和纹理特征,预测原位癌的治疗风险,测试集上准确率在0.7左右;OGURA等[13]通过手工标记出的细胞核,利用机器学习算法,设计和计算细胞核形态特征来预测分析口腔鳞癌的化疗疗效;DIAO等[14]通过对皮肤癌、胃腺癌、肺癌、乳腺癌4种癌症分别提取组织和细胞核层次的手工特征进行分子表型的预测,乳腺癌的分子分型的准确率平均在0.75左右。因此,乳腺癌患者化疗前穿刺切片上肿瘤区域的细胞群层次的特征对于NAC疗效具有预测作用。
乳腺癌病理穿刺切片尺寸较大[15],且组织结构较为复杂,细胞核的种类、形态较多,这些都成为了提取肿瘤区域细胞核层次特征的一大难题。由病理医生进行肿瘤区域和细胞核的标记工作成本较大且不现实,所以,对穿刺切片进行肿瘤区域和细胞核的自动分割是提取肿瘤区域的细胞核层次特征的重要前提。
目前利用提取手工特征定量表征肿瘤区域细胞、构建NAC病理缓解程度的预测模型的工作尚未见有具体结果发表。从病理角度寻找微观的、能够定量分析肿瘤微环境的特征,预测NAC后患者的病理缓解程度,是一个具有重要研究价值的工作。因此,本研究期望利用深度学习方法对乳腺癌患者化疗前的穿刺切片进行肿瘤区域和细胞核的分割,通过提取肿瘤区域细胞群特征来预测Miller-Payne(MP)分级(MP分级越高表示NAC效果越好),借此探索肿瘤区域细胞群特征与患者的NAC疗效之间的关联性。
1. 材料与方法
1.1 实验设备
实验使用的硬件配置如下:处理器:Intel®CoreTM i7-9770 CPU 4 GHz;独立显卡:GeForce GTX 2080Ti;内存:32.0 GB;系统类型:Ubuntu 16.04;开发工具:Python,OpenSlide,Keras框架;读图软件:Qupath。
1.2 实验数据的采集
乳腺癌穿刺切片来自江苏省人民医院的68例患者的NAC前穿刺苏木素-伊红染色(HE)染色切片,纳入患者均在进行NAC后进行了乳腺癌肿瘤切除术并制作了肿瘤全扫描切片,由经验丰富的医生根据对术后全扫描切片的观测得到MP分级,用于描述新辅助化疗的疗效。本研究符合2013年修订的《世界医学会赫尔辛基宣言》,通过了医院伦理委员会批准,批准号2015-SR-220。
两位病理医生对其中12张穿刺切片进行了肿瘤区域的标记,其中8张作为训练集,4张作为测试集,剩余的56张由训练好的肿瘤区分割模型进行肿瘤分割,为下一步工作提供前提条件。细胞核分割模型的训练数据集是基于TCGA公开数据集进行细胞核标记[16]。总共30张1 000×1 000像素的图像块,25张用于训练,5张用于测试。
特征提取利用上述训练好的模型对所有穿刺切片进行了肿瘤区域的分割,计算每个肿瘤区域块中肿瘤区域的占比,选择超过70%的肿瘤区域块进行特征提取。68例患者中在去除几乎没有肿瘤区域穿刺切片后,低MP分级的有45张,高MP级别有23张,采用五折交叉验证,得到模型结果。
1.3 实验步骤
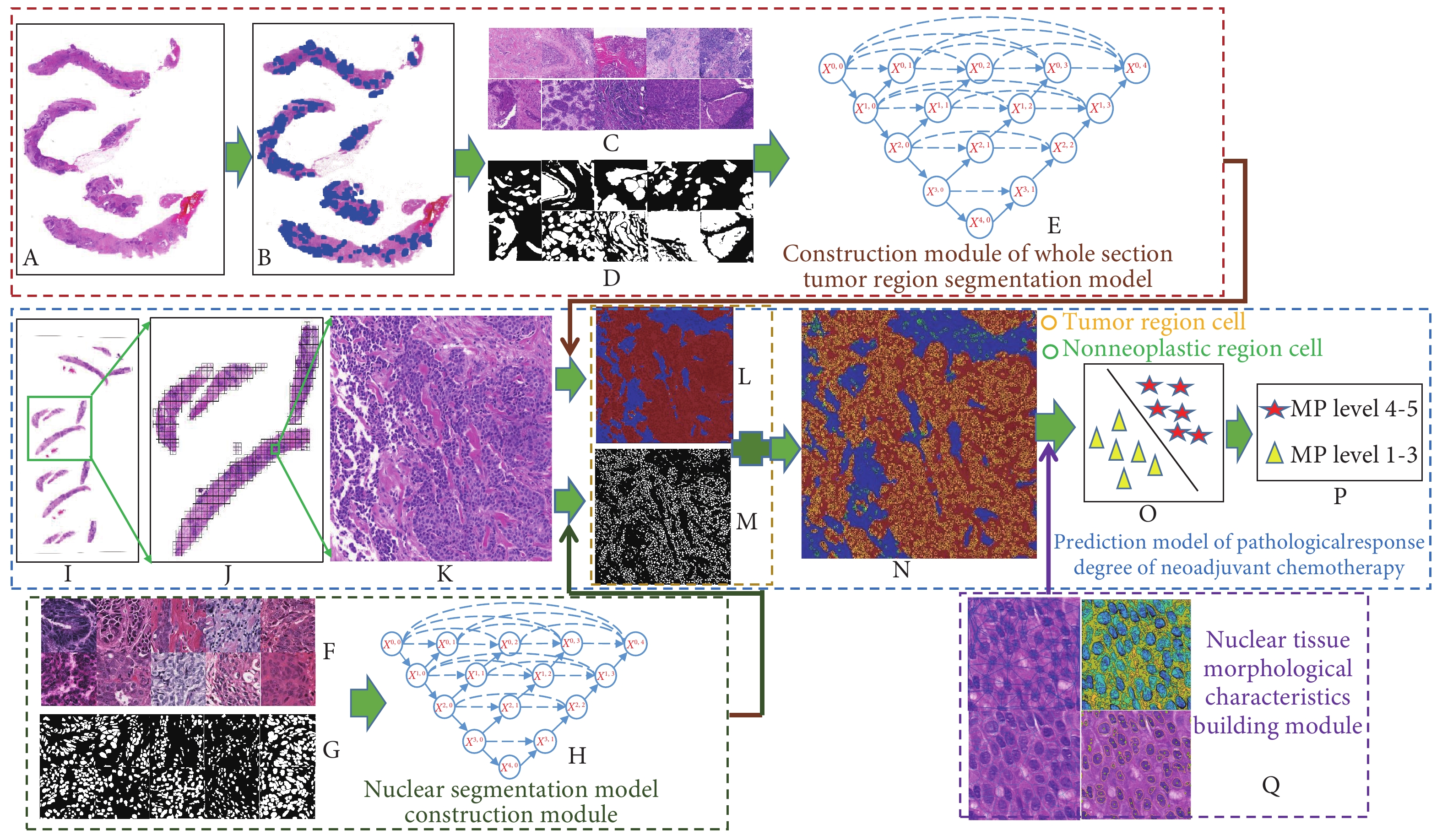
基于乳腺癌患者NAC前的病理穿刺切片的组织形态学对NAC疗效进行预测,预测流程图如图1所示。分为两个模块。①分割训练模块:乳腺肿瘤区域分割(图1A~1E)、细胞核分割(图1F~1H),②预测模块(图1I~1Q):组织形态学特征提取、训练分类器模型。
![]() 图 1 基于组织形态学的乳腺癌NAC疗效的预测流程图Figure 1. The flowchart of treatment outcome prediction of neoadjuvant chemotherapy for breast cancer based on histomorphology analysisA: Needle biopsy image before chemotherapy; B: The marking of needle biopsy image; C: Original patch; D: The marking of patch; E: UNet++; F: Cell cluster segmentation image patch; G: The marking of cell; H: UNet++; I: Needle biopsy image; J: Patch extracted with sliding window; K: Original patch; L: The result of tumor area segmentation; M: The result of cell cluster segmentation; N: The result of segmentation of cell clusters and tumor areas; O: Classifier model; P: The result of prediction of the classifier and grading of response to NAC; Q: Visualization of features of cell clusters in the tumor area.
图 1 基于组织形态学的乳腺癌NAC疗效的预测流程图Figure 1. The flowchart of treatment outcome prediction of neoadjuvant chemotherapy for breast cancer based on histomorphology analysisA: Needle biopsy image before chemotherapy; B: The marking of needle biopsy image; C: Original patch; D: The marking of patch; E: UNet++; F: Cell cluster segmentation image patch; G: The marking of cell; H: UNet++; I: Needle biopsy image; J: Patch extracted with sliding window; K: Original patch; L: The result of tumor area segmentation; M: The result of cell cluster segmentation; N: The result of segmentation of cell clusters and tumor areas; O: Classifier model; P: The result of prediction of the classifier and grading of response to NAC; Q: Visualization of features of cell clusters in the tumor area.1.3.1 分割训练模块的建立
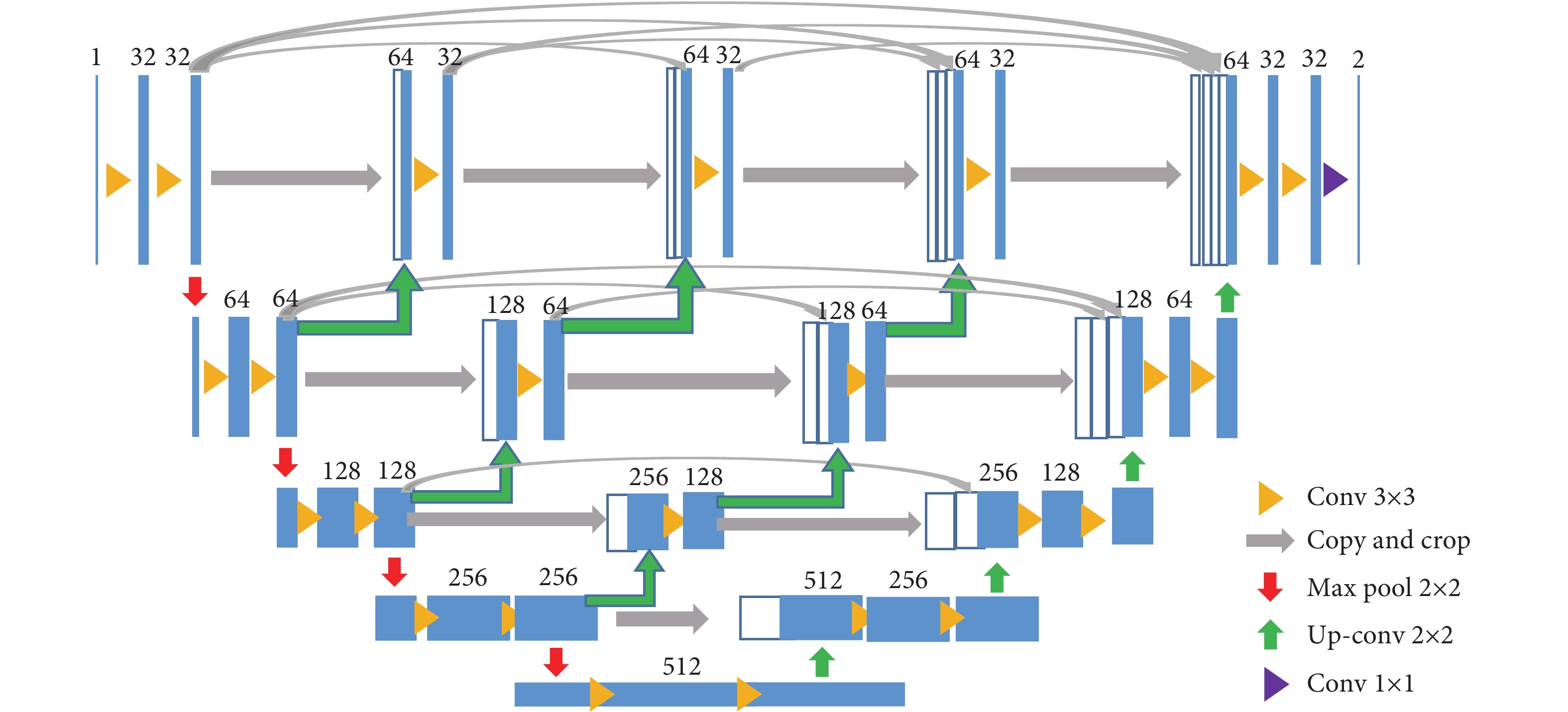
使用语义分割分别建立肿瘤分割和细胞核分割。两个模型使用的网络均是UNet++[17],它能够兼顾图像的浅层和深层的信息,分割的准确率较高。其具体的网络参数结构如图2所示。选择的优化器是自适应矩估计优化器(Adam)[18],能够自适应学习率。模型的损失函数是Dice系数差异函数,通过不断减少Dice系数差异函数来优化网络模型。Dice系数是计算两个样本内的相似度[19]的一种度量函数,如公式(1),公式中这两个样本分别是图像的预测值和真实值。
$$s = \frac{{2|{Y_{{\rm{true}}}} \cap {Y_{{\rm{predict}}}}|}}{{{Y_{{\rm{true}}}} + {Y_{{\rm{predict}}}}}}$$ (1) 其中,
${Y_{{\rm{true}}}}$ 表示训练数据的真实标签值,${Y_{{\rm{predict}}}}$ 表示当前模型正向迭代过程中的预测值。Dice系数差异函数计算如公式(2),通过不断减小公式(2)中的损失函数来优化模型。
$$Loss = 1 - s$$ (2) 1.3.1.1 肿瘤区域分割模块
①滑动窗取块。将整张穿刺切片放大到50倍下取出标记的肿瘤区域,通过滑动窗的方式取出像素为512×512,并将其对应的标记转化成二值图,标记线内的肿瘤区域用白色表示,非肿瘤区域用黑色表示。
②数据筛选。由于肿瘤区域采用的语义分割,其对数据标记的准确度要求较高,而病理图像较大,对整张穿刺切片的标记存在较多遗漏,难以训练一个精确的分割模型。故而将对标记出的穿刺切片通过滑动窗取块的方式分别取出标记完整且图像清晰的块,用于肿瘤分割模型的训练和测试。总共选择了233个标记的完全准确的块,其中对于训练集和测试集的数据分别为163个块和70个块。
③数据增强。由于乳腺癌亚型的多样性和数据染色的差异性导致训练集极不均衡,因此在构建数据集时需要对已有数据进行数据增强,从而防止由于训练数据较少导致的模型过拟合、增加模型的鲁棒性。数据增强主要采用了旋转、对称、颜色变换等方式,由163张图像数据增加到2 320张。
④数据归一化。对经过原始数据增强过后的训练集数据进行归一化处理,以减少扫描图像因染色和扫描等数据采集过程中产生的误差对图像分割产生的影响。
⑤模型训练。将预处理后的数据集和其对应的标记送入UNet++网络中,进行模型训练,得到训练好的模型。
在剩余未标记的穿刺切片的肿瘤区域分割过程中,首先去除背景,提取穿刺切片的感兴趣区域,去除背景区域;然后,在50倍镜下进行滑动窗取块,接着肿瘤区域分割,将每个块都送入训练好的模型中,进行分割,得到同样大小的概率图,每个像素点的值为该点是肿瘤的概率值,将其结果可视化,概率值越接近1(红色)代表其为肿瘤区域的可能性越大,越接近0(蓝色)代表其为肿瘤区域的可能性越小。
1.3.1.2 细胞核分割模块
细胞核分割的训练过程与肿瘤区域分割类似。
由于实验环境的限制,需要将数据集中1 000×1 000像素的图像块重叠地滑动取出4张512×512像素的图像,由25张1 000×1 000像素的图像块变为100张512×512像素的图像块,通过旋转、对称、颜色变换等数据增强的方式将数据扩充为500个图像块,扩充后的数据送入UNet++网络中训练,采取同样的训练机制进行训练。测试集的5张1 000×1 000像素的图像块滑动取出20张512×512的像素块送入训练好的模型中进行测试,选出最好的分割模型。
1.3.2 病理缓解程度预测模块
对68例患者的乳腺癌穿刺切片所选的11 970个512×512像素块的肿瘤分割结果和细胞核分割结果进行叠加,得到肿瘤区域细胞核的分割结果,对肿瘤区域内的细胞核进行细胞核层次的特征提取。
①首先,对所有图像进行了四类特征提取,其中纹理特征具有720维,细胞核全局特征26维,形态特征24维,细胞核聚类特征有24维。总共提取了794维特征。纹理特征的提取是提取每个细胞核中的同质现象,体现其表层结构排列规则。本研究中的纹理特征主要是利用灰度共生矩阵[20] (GLCM)统计细胞核像素的灰度值分布,计算二阶统计量的中位数和方差等。细胞核的全局特征,从整张图像上寻找细胞核的空间分布和拓扑结构。细胞核的形态特征,根据细胞核分割的结果,提取细胞核轮廓特征和区域特征,分别采用了傅里叶形状描述符法[21]、形状无关矩阵计算特征。细胞核聚类特征,寻找到细胞核的群簇质心,计算群簇的空间结构和拓扑关系。表1给出提取的所有特征列表。其中纹理特征共计80维,因为特征的提取是基于单通道的灰度图像进行,所以三通道的RGB图像将会产生240维特征,再计算其均值、方差、熵值,共计720维特征。
表 1 组织形态学特征表Table 1. The list of histomorphological featuresMorphological feature n Texture features 80 Grayscale 15 LBP 16 Gabor 24 Laws 25 Morphological characteristics 24 Frequency domain 10 Distance 7 Invariant moment 7 Global characteristics 24 Delaunay triangle 8 Voronoi 12 Minimum spanning tree 4 Clustering characteristics 26 Basic parameters 6 Clustering coefficient 6 Clustering edge 4 Clustering node 4 Parameters at 90% distance 6 ②将四类特征合并,进行特征排序。特征选择主要利用了4种选择方法,包括最大相关最小冗余(minimal-redundancy-maximum-relevancy approach, mRMR)、秩和检验(Wilcoxon rank-sum test)、Relief特征选择和Fisher精确检验。mRMR[22]是最大化特征与分类标签的相关性,最小化特征与特征的相关性,将所有特征进行排序。Wilcoxon[23]是一种非参数的检验方法,根据不同特征,在不同标签上计算秩和,不同标签上差异越大该特征在区分两类时的作用越大,从而进行特征排序。Relief[24]算法适用于两类样本,通过不断计算特征对 最邻近样本的区分能力以设定每个特征的权重值,根据权重进行排序得到合数特征。Fisher[25]通过计算类内协方差尽可能小、类间离散尽可能大的准则来进行特征排序。
③将4种特征排序的方法选出前10个维度的特征构成特征组合,进行分类器的训练。
④将4种特征选择方法与6种分类器进行两两组合,寻找最优的组合方式。6种分类器分别是随机森林(random forest,RF)、线性判别分析、决策树、逻辑回归、k近邻分类器和支持向量器(support vector machine,SVM)[26]。
⑤每种特征选择方法和每个分类器采用五折交叉验证的方式,得到5个模型,预测出患者每张图像块的MP分级[27](表2),取均值,并与每张图片真实的MP分级进行对比,得到预测模块的准确率以评估模型的预测效果。最终每个患者的预测结果以患者的穿刺切片高MP分级的图像块占选择出的总图像块的比例表示。为了描述方便,本研究中将1、2、3级作为MP分级低,标签为0,病理缓解程度较低;4级和5级为MP分级高,标签为1,病理缓解程度较高。
表 2 MP分级的临床意义Table 2. The clinical meaning of Miller-Payne (MP) systemMP Morphological characteristics of tumor cells Low grades Level 1 No reduction in tumor cell Level 2 No more than 30% reduction of tumor cell Level 3 30%−90% reduction of tumor cell High grades Level 4 More than 90% reduction of tumor cell Level 5 No tumor cell or ductal carcinoma in situ 2. 结果
2.1 肿瘤区域细胞核分割结果
采用滑动窗的方式取块,逐块进行分割,肿瘤区域分割模块最终训练集上的准确率为98.48%,独立测试集上的准确率为92.13%。细胞核分割模块训练集的准确率为99.21%,独立测试集为89.30%。从图3可以发现,本文的分割方法能够较好地分割出肿瘤区域细胞核和肿瘤区域外细胞核,独立测试集上准确率较高,分割效果精确,为进一步利用肿瘤内细胞核来提取细胞核层次的特征,构建有效肿瘤区域细胞群特征来预测NAC后患者病理缓解程度提供了前提条件。
![]() 图 3 肿瘤区域和细胞核分割结果图。 ×50Figure 3. Tumor area and cell segmentation results of whole slide image. ×50A: Original patch; B: The result of tumor segmentation,with red area representing tumor and blue area representing non-tumor; C: The result of cell segmentation, with yellow representing cells in tumor area, and green representing cells out of tumor area.
图 3 肿瘤区域和细胞核分割结果图。 ×50Figure 3. Tumor area and cell segmentation results of whole slide image. ×50A: Original patch; B: The result of tumor segmentation,with red area representing tumor and blue area representing non-tumor; C: The result of cell segmentation, with yellow representing cells in tumor area, and green representing cells out of tumor area.2.2 病理缓解程度预测模块结果与分析
本文主要提取的是肿瘤区域细胞核层次的特征,图4是4种特征的可视化结果展示。将提取到的特征进行特征排序、特征选择,选择出能够有效预测NAC病理缓解程度的特征。
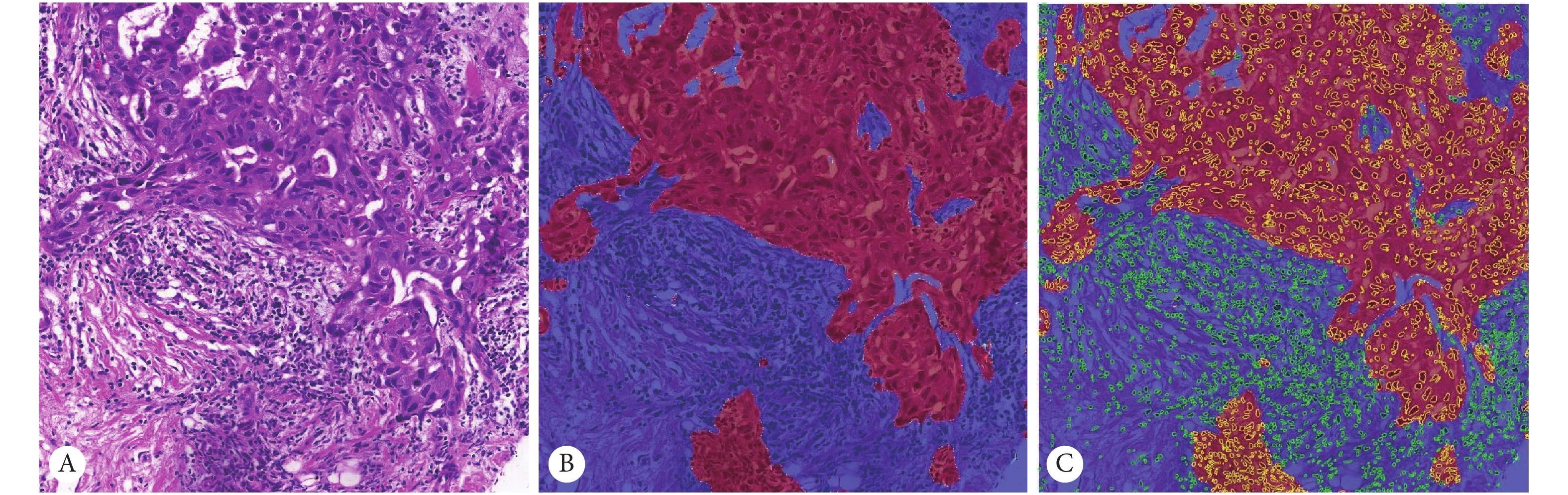
![]() 图 4 特征可视化结果。 ×400Figure 4. The results of feature visualization. ×400A: The display of the Delaunay triangle of cells in tumor area, calculating the distance between the cell centers; B: The area of each tumor cell calculated by the Tyson polygon; C: The visualization of nuclear morphology; D: The visualization of cell texture, showing the texture on the nucleus.
图 4 特征可视化结果。 ×400Figure 4. The results of feature visualization. ×400A: The display of the Delaunay triangle of cells in tumor area, calculating the distance between the cell centers; B: The area of each tumor cell calculated by the Tyson polygon; C: The visualization of nuclear morphology; D: The visualization of cell texture, showing the texture on the nucleus.表3是mRMR、Wilcoxon、Fisher、Relief这4种特征排序方法中前10维特征在选择其最优模型后训练的分类器的结果,准确率(accuracy,ACC)最高达到0.8255,分类效果较好。
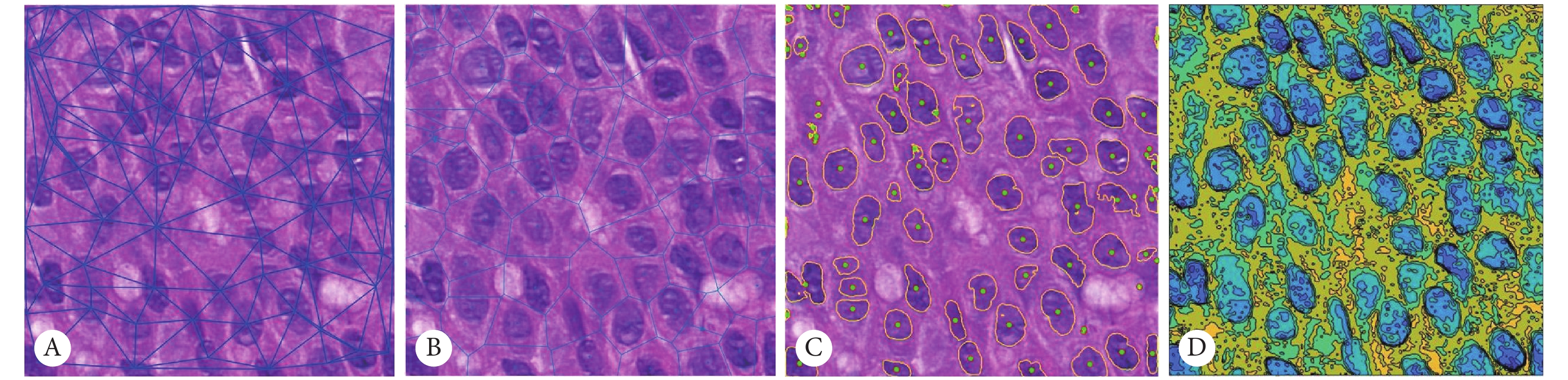
表 3 四种特征排序前10维特征结果Table 3. The results of top ten features in four feature rankingFeature ranking method (10) and classifier 5-fold cross-validation (AUC) 5-fold cross-validation (ACC) mRMR-RF 0.9082 0.8235 Relief-RF 0.8725 0.8235 Fisher-SVM 0.8676 0.7794 Wilcoxon-SVM 0.8174 0.7206 AUC: Area under curve; ACC: Accuracy. 图5是基于受试者工作特征曲线(receiver operating characteristic,ROC)的曲线下面积(area under curve,AUC)的分类结果展示。由图中可以看出mRMR的特征排序方法与RF结合训练出的分类器的五折交叉验证准确率最高,预测效果比其他特征排序的前10维训练出的模型效果好,其ACC为82.35%,AUC达到0.9082。选择出的10维特征均是肿瘤区域细胞核的纹理特征,包括4维Laws特征、3维Gabor特征、2维Grayscale特征和1维LBP特征。这证明了肿瘤区域内部的细胞核特征对于乳腺癌NAC疗效预测的可行性,可以辅助医生指导临床决策。
![]() 图 5 ROC曲线Figure 5. ROC curvesThe red line represents the combination result of ten dimensional feature selected by mRMR and its optimal classifier random forest; the green line represents the combination result of Relief's ten dimensional feature and RF, and the purple line represents the combination result of Fisher and SVM,the blue line represents the combination result of ten dimensional feature selected by Wilcoxon rank-sum test and SVM. AUC: Area under curve; ACC: Accuracy.
图 5 ROC曲线Figure 5. ROC curvesThe red line represents the combination result of ten dimensional feature selected by mRMR and its optimal classifier random forest; the green line represents the combination result of Relief's ten dimensional feature and RF, and the purple line represents the combination result of Fisher and SVM,the blue line represents the combination result of ten dimensional feature selected by Wilcoxon rank-sum test and SVM. AUC: Area under curve; ACC: Accuracy.3. 讨论
本实验的主要目的是通过定量分析乳腺癌患者化疗前的穿刺切片组织形态学特征预测NAC对患者的病理缓解程度的高低。目前研究者主要是通过影像组学进行建模分析NAC的疗效,尚无利用手工特征从病理组学的角度进行NAC的预测。本研究的主要创新点如下:①基于患者的NAC前穿刺病理切片,运用深度网络(UNet++)自动分割肿瘤区域和细胞核,能够准确分割肿瘤区域细胞核,无需手工介入且速度较快,是进行提取肿瘤区域细胞核特征的重要前提。②本文基于上述分割结果,构建组织形态学特征定量描述肿瘤区域细胞核纹理、形态,找到10个维度的特征有效预测NAC病理缓解程度,为医生判断患者是否进行NAC提供一定依据。
本文发现选择出的10个维度的特征均是细胞核层次的纹理特征,对于预测NAC病理缓解至关重要,这与非小细胞肺癌中的细胞核纹理特征对于预后有指导作用[28]一致。纹理特征用于量化感兴趣区域内邻近像素之间的相关性,这证明了在乳腺癌HE染色穿刺切片提取细胞核纹理,预测NAC的病理缓解程度的可行性。这些定量的特征很难通过人肉眼观察发现,但可以被计算机快速、高效地识别。从病理角度寻找微观的、能够定量分析特征,预测NAC后患者的病理缓解程度,更加严谨,模型产生的预测结果可以辅助医生指导临床决策,提高医疗精准度。
文中基于病理穿刺切片,利用深度网络自动分割、提取肿瘤微环境的特征,为NAC病理的研究提供了新的思路,而且效果更好。但仍然存在一些缺陷。由于数据量较少且来自同一家医院,未能分出训练、测试集,且缺乏多中心验证;目前收集到数据的病理缓解程度为5种级别,但是数据量极不均衡,尤其是第五级别的数据较少,所以无法将患者的病理缓解程度分为病理完全缓解和病理非完全缓解,进行精准预测缓解的级别。在未来的研究中可以在多个中心进行扩充数据量,同时增加术后大切片,对化疗前穿刺切片和术后肿瘤切片分别进行肿瘤区域的细胞层次特征的预测和评估,从而进一步验证肿瘤区域细胞群的预测能力,提高乳腺癌疗效预测的精准度。
-
图 1 基于组织形态学的乳腺癌NAC疗效的预测流程图
Figure 1. The flowchart of treatment outcome prediction of neoadjuvant chemotherapy for breast cancer based on histomorphology analysis
A: Needle biopsy image before chemotherapy; B: The marking of needle biopsy image; C: Original patch; D: The marking of patch; E: UNet++; F: Cell cluster segmentation image patch; G: The marking of cell; H: UNet++; I: Needle biopsy image; J: Patch extracted with sliding window; K: Original patch; L: The result of tumor area segmentation; M: The result of cell cluster segmentation; N: The result of segmentation of cell clusters and tumor areas; O: Classifier model; P: The result of prediction of the classifier and grading of response to NAC; Q: Visualization of features of cell clusters in the tumor area.
图 3 肿瘤区域和细胞核分割结果图。 ×50
Figure 3. Tumor area and cell segmentation results of whole slide image. ×50
A: Original patch; B: The result of tumor segmentation,with red area representing tumor and blue area representing non-tumor; C: The result of cell segmentation, with yellow representing cells in tumor area, and green representing cells out of tumor area.
图 4 特征可视化结果。 ×400
Figure 4. The results of feature visualization. ×400
A: The display of the Delaunay triangle of cells in tumor area, calculating the distance between the cell centers; B: The area of each tumor cell calculated by the Tyson polygon; C: The visualization of nuclear morphology; D: The visualization of cell texture, showing the texture on the nucleus.
图 5 ROC曲线
Figure 5. ROC curves
The red line represents the combination result of ten dimensional feature selected by mRMR and its optimal classifier random forest; the green line represents the combination result of Relief's ten dimensional feature and RF, and the purple line represents the combination result of Fisher and SVM,the blue line represents the combination result of ten dimensional feature selected by Wilcoxon rank-sum test and SVM. AUC: Area under curve; ACC: Accuracy.
表 1 组织形态学特征表
Table 1 The list of histomorphological features
Morphological feature n Texture features 80 Grayscale 15 LBP 16 Gabor 24 Laws 25 Morphological characteristics 24 Frequency domain 10 Distance 7 Invariant moment 7 Global characteristics 24 Delaunay triangle 8 Voronoi 12 Minimum spanning tree 4 Clustering characteristics 26 Basic parameters 6 Clustering coefficient 6 Clustering edge 4 Clustering node 4 Parameters at 90% distance 6 表 2 MP分级的临床意义
Table 2 The clinical meaning of Miller-Payne (MP) system
MP Morphological characteristics of tumor cells Low grades Level 1 No reduction in tumor cell Level 2 No more than 30% reduction of tumor cell Level 3 30%−90% reduction of tumor cell High grades Level 4 More than 90% reduction of tumor cell Level 5 No tumor cell or ductal carcinoma in situ 表 3 四种特征排序前10维特征结果
Table 3 The results of top ten features in four feature ranking
Feature ranking method (10) and classifier 5-fold cross-validation (AUC) 5-fold cross-validation (ACC) mRMR-RF 0.9082 0.8235 Relief-RF 0.8725 0.8235 Fisher-SVM 0.8676 0.7794 Wilcoxon-SVM 0.8174 0.7206 AUC: Area under curve; ACC: Accuracy. -
[1] 储春强, 华志元, 刘晓, 等. 乳腺癌术后至化疗的间隔时间对乳腺癌患者治疗效果的影响. 癌症进展,2019,17(24): 2956–2958. DOI: 10.11877/j.issn.1672-1535.2019.17.24.23 [2] DESANTIS C E, MA J, GAUDET M M, et al. Breast cancer statistics, 2019. CA Cancer J Clin,2019,69(6): 438–451. DOI: 10.3322/caac.21583
[3] 朱林林, 王海燕, 颜宪书. 超声造影对浸润性乳腺癌患者的新辅助化疗早期应答的预测价值. 中国药物与临床,2020,20(3): 363–365. DOI: 10.11655/zgywylc2020.03.010 [4] SYMMANS W F, WEI C, GOULD R, et al. Long-term prognostic risk after neoadjuvant chemotherapy associated with residual cancer burden and breast cancer subtype. J Clin Oncol,2017,35(10): 1049–1060. DOI: 10.1200/JCO.2015.63.1010
[5] KRIJGSMAN D, VAN LEEUWEN M, VAN DER VEN J, et al. Quantitative whole slide assessment of tumor-infiltrating CD8-positive lymphocytes in ER-positive breast cancer in relation to clinical outcome. IEEE J Biomed Health Inform,2021,25(2): 381–392. DOI: 10.1109/JBHI.2020.3003475
[6] KUMAR S, BADHE B A, KRISHNAN K M, et al. Study of tumour cellularity in locally advanced breast carcinoma on neo-adjuvant chemotherapy. J Clin Diagn Res,2014,8(4): FC09–13. DOI: 10.7860/JCDR/2014/7594.4283
[7] YAMAGUCHI R, TANAKA M, YANO A, et al. Tumor-infiltrating lymphocytes are important pathologic predictors for neoadjuvant chemotherapy in patients with breast cancer. Human Pathol,2012,43(10): 1688–1694. DOI: 10.1016/j.humpath.2011.12.013
[8] ALI H R, DARIUSH A, PROVENZANO E, et al. Computational pathology of pre-treatment biopsies identifies lymphocyte density as a predictor of response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res,2016,18(1): 21–32. DOI: 10.1186/s13058-016-0682-8
[9] ASANO Y, KASHIWAGI S, GOTO W, et al. Tumour-infiltrating CD8 to FOXP3 lymphocyte ratio in predicting treatment responses to neoadjuvant chemotherapy of aggressive breast cancer. Brit J Surg,2016,103(7): 845–854. DOI: 10.1002/bjs.10127
[10] GARCÍA-MARTÍNEZ E, GIL G L, BENITO A C, et al. Tumor-infiltrating immune cell profiles and their change after neoadjuvant chemotherapy predict response and prognosis of breast cancer. Breast Cancer Res,2014,16(6): 488. DOI: 10.1186/s13058-014-0488-5
[11] NISHIMURA R, OSAKO T, OKUMURA Y, et al. Clinical significance of Ki-67 in neoadjuvant chemotherapy for primary breast cancer as a predictor for chemosensitivity and for prognosis. Breast Cancer,2010,17(4): 269–275. DOI: 10.1007/s12282-009-0161-5
[12] LI H J, WHITNEY J, BERA K, et al. Quantitative nuclear histomorphometric features are predictive of oncotype DX risk categories in ductal carcinoma in situ: preliminary findings. Breast Cancer Res,2019,21(1): 114–130. DOI: 10.1186/s13058-019-1200-6
[13] OGURA M, YAMAMOTO Y, MIYASHITA H, et al. Quantitative analysis of nuclear shape in oral squamous cell carcinoma is useful for predicting the chemotherapeutic response. Med Mol Morphol,2016,49(2): 76–82. DOI: 10.1007/s00795-015-0121-4
[14] DIAO J A, CHUI W F, WANG J K, et al. Dense, high-resolution mapping of cells and tissues from pathology images for the interpretable prediction of molecular phenotypes in cancer. bioRxiv, 2020[2020-10-08]. doi: 10.1101/2020.08.02.233197.
[15] LI J, LI W, SISK A, et al. A multi-resolution model for histopathology image classification and localization with multiple instance learning. Comput Biol Med, 2021, 131: 104253[2020-10-08]. https://doi.org/10.1016/j.compbiomed.2021.104253.
[16] KUMAR N, VERMA R, SHARMA S, et al. A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans Med Imaging,2017,36(7): 1550–1560. DOI: 10.1109/TMI.2017.2677499
[17] ZHOU Z, SIDDIQUEE M M R, TAJBAKHSH N, et al. Unet++: a nested u-net architecture for medical image segmentation//Deep learning in medical image analysis and multimodal learning for clinical decision support. Cham: Springer, 2018: 3-11.
[18] KINGMA D P, BA J. Adam: a method for stochastic optimization//3rd International Conference for Learning Representations. San Diego.2014: 5-18[2020-10-08]. https://arxiv.org/abs/1412.6980.
[19] ANUAR N, SULTAN A B M. Validate conference paper using dice coefficient. Computer Informat Sci,2010,3(3): 139–145.
[20] MOHANAIAH P, SATHYANARAYANA P, GURUKUMAR L. Image texture feature extraction using GLCM approach. Int J Scientific Res Publication,2013,3(5): 1–5.
[21] DUYCKAERTS C, GODEFROY G. Voronoi tessellation to study the numerical density and the spatial distribution of neurones. J Chem Neuroanatomy,2000,20(1): 83–92. DOI: 10.1016/S0891-0618(00)00064-8
[22] PENG H, LONG F, DING C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell,2005,27(8): 1226–1238. DOI: 10.1109/TPAMI.2005.159
[23] CUZICK J. A Wilcoxon‐type test for trend. Statist Med,1985,4(1): 87–90. DOI: 10.1002/sim.4780040112
[24] KONONENKO I. Estimating attributes: analysis and extensions of RELIEF//European conference on machine learning. Berlin, Heidelberg: Springer, 1994: 171-182.
[25] MIKA S, RATSCH G, WESTON J, et al. Fisher discriminant analysis with kernels//Neural networks for signal processing Ⅸ: Proceedings of the 1999 IEEE signal processing society workshop (cat. no. 98th8468). IEEE, 1999: 41-48.
[26] TONG S, KOLLER D. Support vector machine active learning with applications to text classification. J Machine Learn Res,2001,2(11): 45–66. DOI: 10.1162/153244302760185243
[27] 单慧明, 周靖宇, 谢婷婷, 等. MRI影像学特征预测乳腺癌新辅助化疗疗效的可行性. 中国医学影像学杂志,2019,27(12): 905–909. DOI: 10.3969/j.issn.1005-5185.2019.12.006 [28] YU KH, ZHANG C, BERRY G J, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun, 2016, 7: 12474[2020-10-08]. https://www.nature.com/articles/ncomms12474. doi: 10.1038/ncomms12474.
-
期刊类型引用(2)
1. 王亚敏. 海曲泊帕乙醇胺片联合rhTPO治疗实体瘤化疗后血小板减少症患者的临床研究. 中国医学创新. 2024(14): 65-68 .  百度学术
百度学术
2. 康宁,王宇立,方志红. 基于“诸寒收引,皆属于肾”探讨从肾论治胃癌化学疗法后骨髓抑制. 上海中医药杂志. 2023(11): 17-21 .  百度学术
百度学术
其他类型引用(3)

 首页
首页


 下载:
下载: